Overview
In this tutorial, you will:- Create a dataset in Labelbox
- Import custom metadata and ground truth
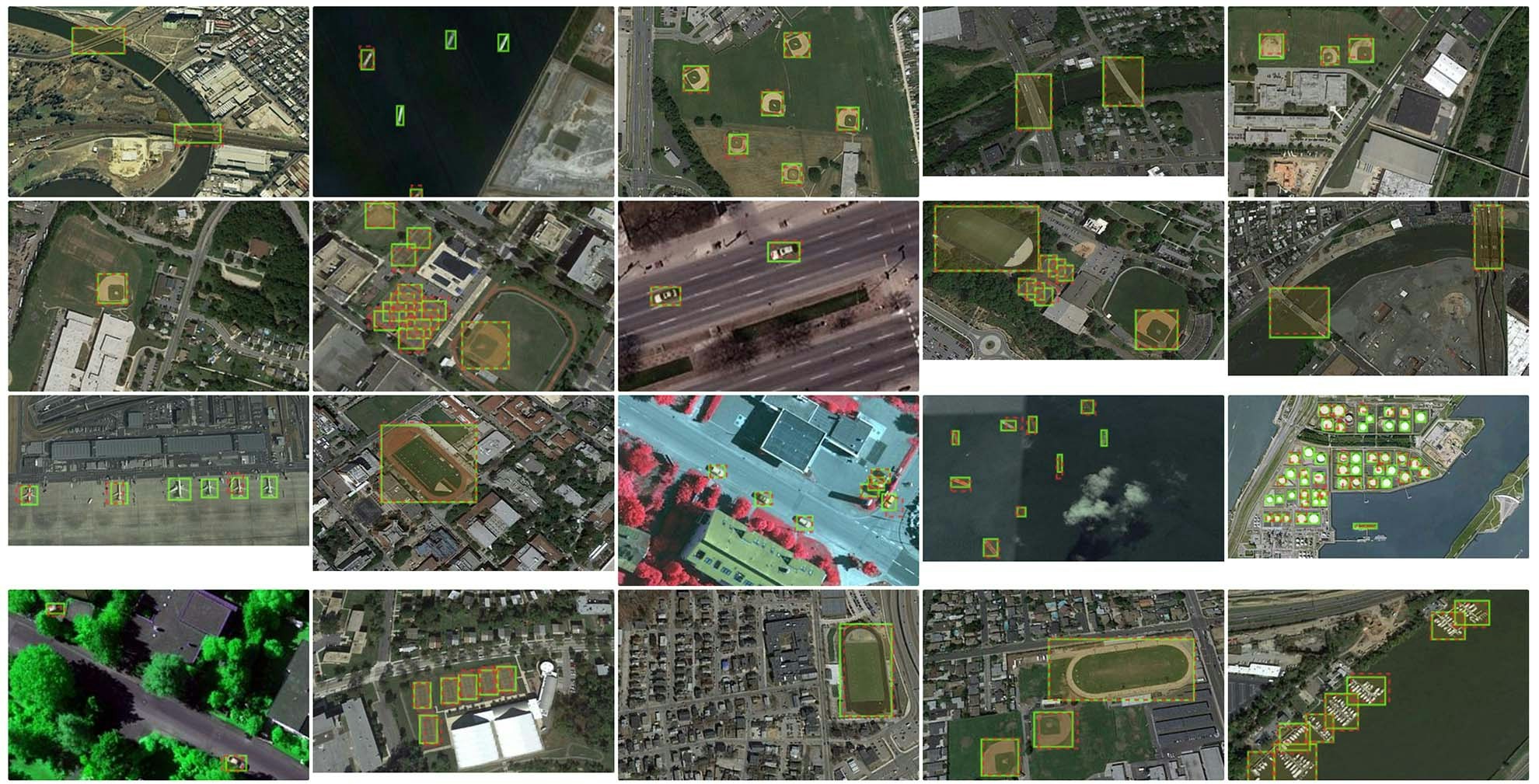
End-to-end example: import ground truth
Import libraries
Copy
Ask AI
import labelbox as lb
from labelbox.schema.data_row_metadata import DataRowMetadataField, DataRowMetadataKind
import datetime
import random
import os
import json
from PIL import Image
from labelbox.schema.ontology import OntologyBuilder, Tool
import requests
from tqdm.notebook import tqdm
import uuid
from labelbox.data.annotation_types import Label, ImageData, ObjectAnnotation, Rectangle
Set up Labelbox client
Copy
Ask AI
# Initialize the Labelbox client
API_KEY = "" # Place API key
client = lb.Client(API_KEY)
Download a public dataset
Copy
Ask AI
# Function to download files
def download_files(filemap):
path, uri = filemap
if not os.path.exists(path):
response = requests.get(uri, stream=True)
with open(path, 'wb') as f:
for chunk in response.iter_content(chunk_size=8192):
f.write(chunk)
return path
Download data rows and annotations
Copy
Ask AI
# Download data rows and annotations
DATA_ROWS_URL = "https://storage.googleapis.com/labelbox-datasets/VHR_geospatial/geospatial_datarows.json"
ANNOTATIONS_URL = "https://storage.googleapis.com/labelbox-datasets/VHR_geospatial/geospatial_annotations.json"
download_files(("data_rows.json", DATA_ROWS_URL))
download_files(("annotations.json", ANNOTATIONS_URL))
Copy
Ask AI
with open('data_rows.json', 'r') as fp:
data_rows = json.load(fp)
with open('annotations.json', 'r') as fp:
annotations = json.load(fp)
Create a dataset
Copy
Ask AI
# Create a new dataset
dataset = client.create_dataset(name="Geospatial vessel detection")
print(f"Created dataset with ID: {dataset.uid}")
Import Data Rows and metadata
Copy
Ask AI
# Here is an example of adding two metadata fields to your Data Rows: a "captureDateTime" field with datetime value, and a "tag" field with string value
metadata_ontology = client.get_data_row_metadata_ontology()
datetime_schema_id = metadata_ontology.reserved_by_name["captureDateTime"].uid
tag_schema_id = metadata_ontology.reserved_by_name["tag"].uid
tag_items = ["WorldView-1", "WorldView-2", "WorldView-3", "WorldView-4"]
for datarow in tqdm(data_rows):
dt = datetime.datetime.utcnow() + datetime.timedelta(days=random.random()*30) # this is random datetime value
tag_item = random.choice(tag_items) # this is a random tag value
# Option 1: Specify metadata with a list of DataRowMetadataField. This is the recommended option since it comes with validation for metadata fields.
metadata_fields = [
DataRowMetadataField(schema_id=datetime_schema_id, value=dt),
DataRowMetadataField(schema_id=tag_schema_id, value=tag_item)
]
# Option 2: Uncomment to try. Alternatively, you can specify the metadata fields with dictionary format without declaring the DataRowMetadataField objects. It is equivalent to Option 1.
# metadata_fields = [
# {"schema_id": datetime_schema_id, "value": dt},
# {"schema_id": tag_schema_id, "value": tag_item}
# ]
datarow["metadata_fields"] = metadata_fields
Copy
Ask AI
task = dataset.create_data_rows(data_rows)
task.wait_till_done()
print(f"Failed data rows: {task.failed_data_rows}")
print(f"Errors: {task.errors}")
if task.errors:
for error in task.errors:
if 'Duplicate global key' in error['message'] and dataset.row_count == 0:
# If the global key already exists in the workspace the dataset will be created empty, so we can delete it.
print(f"Deleting empty dataset: {dataset}")
dataset.delete()
Examine a data row
Copy
Ask AI
datarow = next(dataset.data_rows())
print(datarow)
Set up a labeling project
Copy
Ask AI
# Initialize the OntologyBuilder
ontology_builder = OntologyBuilder()
# Assuming 'annotations' is defined and contains the necessary data
for category in annotations['categories']:
print(category['name'])
# Add tools to the ontology builder
ontology_builder.add_tool(Tool(tool=Tool.Type.BBOX, name=category['name']))
# Create the ontology in Labelbox
ontology = client.create_ontology("Vessel Detection Ontology",
ontology_builder.asdict(),
media_type=lb.MediaType.Image)
print(f"Created ontology with ID: {ontology.uid}")
# Create a project and connect the ontology
project = client.create_project(name="Vessel Detection", media_type=lb.MediaType.Image)
project.connect_ontology(ontology=ontology)
print(f"Created project with ID: {project.uid}")
Send a batch of data rows to the project
Copy
Ask AI
# Minimal Export parameters focused solely on data row IDs
export_params = {
"data_row_details": True # Only export data row details
}
# Initiate the streamable export task from catalog
dataset = client.get_dataset(dataset.uid) # Update with the actual dataset ID
export_task = dataset.export(params=export_params)
export_task.wait_till_done()
print(export_task)
data_rows = []
# Stream the export using a callback function
def json_stream_handler(output: labelbox.BufferedJsonConverterOutput):
print(output.json)
export_task.get_buffered_stream(stream_type=labelbox.StreamType.RESULT).start(stream_handler=json_stream_handler)
# Collect all exported data into a list
export_json = [data_row.json for data_row in export_task.get_buffered_stream()]
# Randomly select 200 Data Rows (or fewer if the dataset has less than 200 data rows)
sampled_data_rows = random.sample(data_rows, min(len(data_rows), 200))
# Create a new batch in the project and add the sampled data rows
batch = project.create_batch(
"Initial batch", # name of the batch
sampled_data_rows, # list of Data Rows
1 # priority between 1-5
)
print(f"Created batch with ID: {batch.uid}")
Create annotations payload
Copy
Ask AI
# Set export parameters focused on data row details
export_params = {
"data_row_details": True, # Only export data row details
"batch_ids": [batch.uid], # Optional: Include batch ids to filter by specific batches
}
# Initialize the streamable export task from project
export_task = project.export(params=export_params)
export_task.wait_till_done()
data_rows = []
# Stream the export using a callback function
def json_stream_handler(output: labelbox.BufferedJsonConverterOutput):
print(output.json)
export_task.get_buffered_stream(stream_type=labelbox.StreamType.RESULT).start(stream_handler=json_stream_handler)
# Collect all exported data into a list
export_json = [data_row.json for data_row in export_task.get_buffered_stream()]
labels = []
for datarow in data_rows:
annotations_list = []
# Access the 'data_row' dictionary first
data_row_dict = datarow['data_row']
folder = data_row_dict['external_id'].split("/")[0]
id = data_row_dict['external_id'].split("/")[1]
if folder == "positive_image_set":
for image in annotations['images']:
if image['file_name'] == id:
for annotation in annotations['annotations']:
if annotation['image_id'] == image['id']:
bbox = annotation['bbox']
category_id = annotation['category_id'] - 1
class_name = None
ontology = ontology_builder.asdict() # Get the ontology dictionary
for category in ontology['tools']:
if category['name'] == annotations['categories'][category_id]['name']:
class_name = category['name']
break
if class_name:
annotations_list.append(ObjectAnnotation(
name=class_name,
value=Rectangle(start=Point(x=bbox[0], y=bbox[1]), end=Point(x=bbox[2]+bbox[0], y=bbox[3]+bbox[1]))
))
image_data = ImageData(uid=data_row_dict['id'])
labels.append(Label(data=image_data, annotations=annotations_list))
Import ground truth annotations
Copy
Ask AI
upload_job = lb.LabelImport.create_from_objects(
client=client,
project_id=project.uid,
name=f"label_import_job_{str(uuid.uuid4())}",
labels=labels
)
# Wait for the upload to finish and print the results
upload_job.wait_until_done()
print(f"Errors: {upload_job.errors}")
print(f"Status of uploads: {upload_job.statuses}")